Tutorial 08: Data Extraction#
Many post-processing activities require a non-proprietary serialization format to provide common access to a mix of analysis libraries and tools. The Parameter Generators of this project uses the xarray dataset and H5 files, so it will be convenient to extract data results to a similar H5 file using matching coordinates for the N-dimensional data.
Abaqus provides a scripting interface and a proprietary, binary output database file containing the simulation results.
While this object and interface is good for scripting with the Abaqus kernel and CAE features, the Python interface is
limited to the Python 2.7 environment that is shipped with Abaqus. When post-processing requires more Python libraries
or other third party tools, it is common to extract a portion of the Abaqus results to an intermediate file. To reduce
duplicated effort in extracting Abaqus ODB files to a common format, the ODB Extract command-line utility and
the associated waves.scons_extensions.abaqus_extract() builder are provided to parse the output of abaqus odbreport
into several xarray dataset objects in an H5 file.
Future releases of WAVES may include extract utilities for other numeric solvers.
References#
Environment#
SCons and WAVES can be installed in a Conda environment with the Conda package manager. See the Conda installation and Conda environment management documentation for more details about using Conda.
Note
The SALib and numpy versions may not need to be this strict for most tutorials. However, Tutorial: Sensitivity Study uncovered some undocumented SALib version sensitivity to numpy surrounding the numpy v2 rollout.
Create the tutorials environment if it doesn’t exist
$ conda create --name waves-tutorial-env --channel conda-forge waves 'scons>=4.6' matplotlib pandas pyyaml xarray seaborn 'numpy>=2' 'salib>=1.5.1' pytest
PS > conda create --name waves-tutorial-env --channel conda-forge waves scons matplotlib pandas pyyaml xarray seaborn numpy salib pytest
Activate the environment
$ conda activate waves-tutorial-env
PS > conda activate waves-tutorial-env
Some tutorials require additional third-party software that is not available for the Conda package manager. This
software must be installed separately and either made available to SConstruct by modifying your system’s PATH or by
modifying the SConstruct search paths provided to the waves.scons_extensions.add_program() method.
Warning
STOP! Before continuing, check that the documentation version matches your installed package version.
You can find the documentation version in the upper-left corner of the webpage.
You can find the installed WAVES version with
waves --version.
If they don’t match, you can launch identically matched documentation with the WAVES Command-Line Utility
docs subcommand as waves docs.
Directory Structure#
Create and change to a new project root directory to house the tutorial files if you have not already done so. For example
$ mkdir -p ~/waves-tutorials
$ cd ~/waves-tutorials
$ pwd
/home/roppenheimer/waves-tutorials
PS > New-Item $HOME\waves-tutorials -ItemType "Directory"
PS > Set-Location $HOME\waves-tutorials
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
Note
If you skipped any of the previous tutorials, run the following commands to create a copy of the necessary tutorial files.
$ pwd
/home/roppenheimer/waves-tutorials
$ waves fetch --overwrite --tutorial 7 && mv tutorial_07_cartesian_product_SConstruct SConstruct
WAVES fetch
Destination directory: '/home/roppenheimer/waves-tutorials'
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > waves fetch --overwrite --tutorial 7 && Move-Item tutorial_07_cartesian_product_SConstruct SConstruct -Force
WAVES fetch
Destination directory: 'C:\Users\roppenheimer\waves-tutorials'
Download and copy the
tutorial_07_cartesian_product.sconsfile to a new file namedtutorial_08_data_extraction.sconswith the WAVES Command-Line Utility fetch subcommand.
$ pwd
/home/roppenheimer/waves-tutorials
$ waves fetch --overwrite tutorials/tutorial_07_cartesian_product.scons && cp tutorial_07_cartesian_product.scons tutorial_08_data_extraction.scons
WAVES fetch
Destination directory: '/home/roppenheimer/waves-tutorials'
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > waves fetch --overwrite tutorials\tutorial_07_cartesian_product.scons && Copy-Item tutorial_07_cartesian_product.scons tutorial_08_data_extraction.scons
WAVES fetch
Destination directory: 'C:\Users\roppenheimer\waves-tutorials'
SConscript#
A diff against the tutorial_07_cartesian_product.scons file from Tutorial 07: Cartesian Product is included
below to help identify the changes made in this tutorial.
waves-tutorials/tutorial_08_data_extraction.scons
--- /home/runner/work/waves/waves/build/docs/tutorials_tutorial_07_cartesian_product.scons
+++ /home/runner/work/waves/waves/build/docs/tutorials_tutorial_08_data_extraction.scons
@@ -149,6 +149,15 @@
)
)
+ # Extract Abaqus
+ extract_source_list = [set_path / "rectangle_compression.odb"]
+ workflow.extend(
+ env.AbaqusExtract(
+ target=[set_path / "rectangle_compression.h5"],
+ source=extract_source_list,
+ )
+ )
+
# Comment used in tutorial code snippets: marker-6
# Collector alias based on parent directory name
The only new code in this tutorial adds the waves.scons_extensions.abaqus_extract() builder task. Note that this task
falls within the parameterization loop and will be executed once per parameter set. ODB Extract will output
two files: rectangle_compression_datasets.h5, which contains h5py paths to the Xarray datasets, and
rectangle_compression.h5, which contains h5py native datasets for anything that ODB Extract doesn’t
organize into Xarray datasets and a list of group paths pointing at Xarray datasets in
rectangle_compression_datasets.h5. The second file also contains external links to the datasets file, so h5py
can be used to access all group paths if necessary. Tutorial 09: Post-Processing will introduce an example for
accessing and organizing the results files and concatenating the parameter study information.
SConstruct#
Update the
SConstructfile with the changes below.Attach the
waves.scons_extensions.abaqus_extract()builder to the construction environment asAbaqusExtractby appending theBUILDERSdictionary.Add
tutorial_08_data_extractionto the workflow_configurations list.
A diff against the SConstruct file from Tutorial 07: Cartesian Product is included below to help identify the
changes made in this tutorial.
waves-tutorials/SConstruct
--- /home/runner/work/waves/waves/build/docs/tutorials_tutorial_07_cartesian_product_SConstruct
+++ /home/runner/work/waves/waves/build/docs/tutorials_tutorial_08_data_extraction_SConstruct
@@ -1,5 +1,5 @@
#! /usr/bin/env python
-"""Configure the WAVES parameter study tutorial using the cartesian product parameter generator."""
+"""Configure the WAVES data extraction tutorial."""
import os
import pathlib
@@ -93,7 +93,11 @@
# Comments used in tutorial code snippets: marker-5
# Add builders and pseudo-builders
-env.Append(BUILDERS={})
+env.Append(
+ BUILDERS={
+ "AbaqusExtract": waves.scons_extensions.abaqus_extract(program=env["ABAQUS_PROGRAM"]),
+ }
+)
# Comments used in tutorial code snippets: marker-6
@@ -106,6 +110,7 @@
"tutorial_05_parameter_substitution.scons",
"tutorial_06_include_files.scons",
"tutorial_07_cartesian_product.scons",
+ "tutorial_08_data_extraction.scons",
]
for workflow in workflow_configurations:
build_dir = env["variant_dir_base"] / pathlib.Path(workflow).stem
Build Targets#
Build the new targets
$ pwd
/home/roppenheimer/waves-tutorials
$ scons tutorial_08_data_extraction --jobs=4
<output truncated>
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > scons tutorial_08_data_extraction --jobs=4
<output truncated>
Output Files#
View the output files. The output files should match those introduced in Tutorial 07: Cartesian Product, with the addition of the ODB Extract output files.
$ pwd
/home/roppenheimer/waves-tutorials
$ find build/tutorial_08_data_extraction/ -name "*.h5"
build/tutorial_08_data_extraction/parameter_study.h5
build/tutorial_08_data_extraction/parameter_set2/rectangle_compression.h5
build/tutorial_08_data_extraction/parameter_set2/rectangle_compression_datasets.h5
build/tutorial_08_data_extraction/parameter_set1/rectangle_compression.h5
build/tutorial_08_data_extraction/parameter_set1/rectangle_compression_datasets.h5
build/tutorial_08_data_extraction/parameter_set3/rectangle_compression.h5
build/tutorial_08_data_extraction/parameter_set3/rectangle_compression_datasets.h5
build/tutorial_08_data_extraction/parameter_set0/rectangle_compression.h5
build/tutorial_08_data_extraction/parameter_set0/rectangle_compression_datasets.h5
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > Get-ChildItem -Path build\tutorial_08_data_extraction\ -Recurse "*.h5"
Directory: C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 9942 parameter_study.h5
Directory: C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set0
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 48648 rectangle_compression_datasets.h5
-a--- 6/9/2023 4:32 PM 140868 rectangle_compression.h5
Directory: C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set1
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 48648 rectangle_compression_datasets.h5
-a--- 6/9/2023 4:32 PM 140868 rectangle_compression.h5
Directory: C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set2
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 48648 rectangle_compression_datasets.h5
-a--- 6/9/2023 4:32 PM 140868 rectangle_compression.h5
Directory: C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set3
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 48648 rectangle_compression_datasets.h5
-a--- 6/9/2023 4:32 PM 140868 rectangle_compression.h5
See the ODB Extract and waves.scons_extensions.abaqus_extract() documentation for an overview of the H5
file structure. You will notice that there are two H5 files per parameter set. The data is organized into a root file,
rectangle_compression.h5, that contains the overall file structure, some unsorted meta data, and a list of H5 group
paths containing Xarray datasets. The second file, rectangle_compression_datasets.h5, contains the actual Xarray
dataset objects. Tutorial 09: Post-Processing will introduce a post-processing task with the
waves.scons_extensions.abaqus_extract() H5 output files.
You can explore the structure of each file with The HDF5 Group command-line-tools below (the grep command
excludes the large, unsorted data in the /odb group path).
$ h5ls -r build/tutorial_08_data_extraction/parameter_set0/rectangle_compression.h5 | grep -v odb
/ Group
/RECTANGLE Group
/RECTANGLE/FieldOutputs Group
/RECTANGLE/FieldOutputs/ALL_ELEMENTS External Link {/home/roppenheimer/waves-tutorials/build/tutorial_08_data_extraction/parameter_set0/rectangle_compression_datasets.h5//RECTANGLE/FieldOutputs/ALL_ELEMENTS}
/RECTANGLE/FieldOutputs/ALL_NODES External Link {/home/roppenheimer/waves-tutorials/build/tutorial_08_data_extraction/parameter_set0/rectangle_compression_datasets.h5//RECTANGLE/FieldOutputs/ALL_NODES}
/RECTANGLE/HistoryOutputs Group
/RECTANGLE/HistoryOutputs/NODES External Link {/home/roppenheimer/waves-tutorials/build/tutorial_08_data_extraction/parameter_set0/rectangle_compression_datasets.h5//RECTANGLE/HistoryOutputs/NODES}
/RECTANGLE/Mesh External Link {/home/roppenheimer/waves-tutorials/build/tutorial_08_data_extraction/parameter_set0/rectangle_compression_datasets.h5//RECTANGLE/Mesh}
/xarray Group
/xarray/Dataset Dataset {4}
PS > h5ls -r build\tutorial_08_data_extraction\parameter_set0\rectangle_compression.h5 | Where-Object { $_ -notmatch "odb" }
/ Group
/RECTANGLE Group
/RECTANGLE/FieldOutputs Group
/RECTANGLE/FieldOutputs/ALL_ELEMENTS External Link {C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set0\rectangle_compression_datasets.h5//RECTANGLE/FieldOutputs/ALL_ELEMENTS}
/RECTANGLE/FieldOutputs/ALL_NODES External Link {C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set0\rectangle_compression_datasets.h5//RECTANGLE/FieldOutputs/ALL_NODES}
/RECTANGLE/HistoryOutputs Group
/RECTANGLE/HistoryOutputs/NODES External Link {C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set0\rectangle_compression_datasets.h5//RECTANGLE/HistoryOutputs/NODES}
/RECTANGLE/Mesh External Link {C:\Users\roppenheimer\waves-tutorials\build\tutorial_08_data_extraction\parameter_set0\rectangle_compression_datasets.h5//RECTANGLE/Mesh}
/xarray Group
/xarray/Dataset Dataset {4}
$ h5ls -r build/tutorial_08_data_extraction/parameter_set0/rectangle_compression_datasets.h5
/ Group
/RECTANGLE Group
/RECTANGLE/FieldOutputs Group
/RECTANGLE/FieldOutputs/ALL_ELEMENTS Group
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/E Dataset {1, 5, 1, 4, 4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/E\ values Dataset {4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/S Dataset {1, 5, 1, 4, 4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/S\ values Dataset {4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/elements Dataset {1}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/integration\ point Dataset {4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/integrationPoint Dataset {1, 4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/step Dataset {1}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/time Dataset {5}
/RECTANGLE/FieldOutputs/ALL_NODES Group
/RECTANGLE/FieldOutputs/ALL_NODES/U Dataset {1, 5, 4, 2}
/RECTANGLE/FieldOutputs/ALL_NODES/U\ values Dataset {2}
/RECTANGLE/FieldOutputs/ALL_NODES/nodes Dataset {4}
/RECTANGLE/FieldOutputs/ALL_NODES/step Dataset {1}
/RECTANGLE/FieldOutputs/ALL_NODES/time Dataset {5}
/RECTANGLE/HistoryOutputs Group
/RECTANGLE/HistoryOutputs/NODES Group
/RECTANGLE/HistoryOutputs/NODES/U1 Dataset {1, 14}
/RECTANGLE/HistoryOutputs/NODES/U2 Dataset {1, 14}
/RECTANGLE/HistoryOutputs/NODES/node Dataset {1}
/RECTANGLE/HistoryOutputs/NODES/step Dataset {1}
/RECTANGLE/HistoryOutputs/NODES/time Dataset {14}
/RECTANGLE/HistoryOutputs/NODES/type Dataset {1}
/RECTANGLE/Mesh Group
/RECTANGLE/Mesh/CPS4R Dataset {1}
/RECTANGLE/Mesh/CPS4R_mesh Dataset {1, 4}
/RECTANGLE/Mesh/CPS4R_node Dataset {4}
/RECTANGLE/Mesh/node Dataset {4}
/RECTANGLE/Mesh/node_location Dataset {4, 3}
/RECTANGLE/Mesh/section_category Dataset {1}
/RECTANGLE/Mesh/vector Dataset {3}
PS > h5ls -r build\tutorial_08_data_extraction\parameter_set0\rectangle_compression_datasets.h5
/ Group
/RECTANGLE Group
/RECTANGLE/FieldOutputs Group
/RECTANGLE/FieldOutputs/ALL_ELEMENTS Group
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/E Dataset {1, 5, 1, 4, 4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/E\ values Dataset {4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/S Dataset {1, 5, 1, 4, 4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/S\ values Dataset {4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/elements Dataset {1}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/integration\ point Dataset {4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/integrationPoint Dataset {1, 4}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/step Dataset {1}
/RECTANGLE/FieldOutputs/ALL_ELEMENTS/time Dataset {5}
/RECTANGLE/FieldOutputs/ALL_NODES Group
/RECTANGLE/FieldOutputs/ALL_NODES/U Dataset {1, 5, 4, 2}
/RECTANGLE/FieldOutputs/ALL_NODES/U\ values Dataset {2}
/RECTANGLE/FieldOutputs/ALL_NODES/nodes Dataset {4}
/RECTANGLE/FieldOutputs/ALL_NODES/step Dataset {1}
/RECTANGLE/FieldOutputs/ALL_NODES/time Dataset {5}
/RECTANGLE/HistoryOutputs Group
/RECTANGLE/HistoryOutputs/NODES Group
/RECTANGLE/HistoryOutputs/NODES/U1 Dataset {1, 14}
/RECTANGLE/HistoryOutputs/NODES/U2 Dataset {1, 14}
/RECTANGLE/HistoryOutputs/NODES/node Dataset {1}
/RECTANGLE/HistoryOutputs/NODES/step Dataset {1}
/RECTANGLE/HistoryOutputs/NODES/time Dataset {14}
/RECTANGLE/HistoryOutputs/NODES/type Dataset {1}
/RECTANGLE/Mesh Group
/RECTANGLE/Mesh/CPS4R Dataset {1}
/RECTANGLE/Mesh/CPS4R_mesh Dataset {1, 4}
/RECTANGLE/Mesh/CPS4R_node Dataset {4}
/RECTANGLE/Mesh/node Dataset {4}
/RECTANGLE/Mesh/node_location Dataset {4, 3}
/RECTANGLE/Mesh/section_category Dataset {1}
/RECTANGLE/Mesh/vector Dataset {3}
Each group path directly above a Dataset path entry can be opened with Xarray, e.g.
/RECTANGLE/FieldOutputs/ALL_ELEMENTS as
import xarray
extracted_file = "build/tutorial_08_data_extraction/parameter_set0/rectangle_compression.h5"
group_path = "/RECTANGLE/FieldOutputs/ALL_ELEMENTS"
field_outputs = xarray.open_dataset(extracted_file, group=group_path)
The structure is separated into two files to aid automated exploration of the data structure. The root file,
rectangle_compression.h5 may be opened with h5py to obtain the Xarray datasets list in the /xarray/Dataset
group path. Then the datasets file, rectangle_compression_datasets.h5, may be opened separately with Xarray in
the same Python script. This is required because h5py and Xarray can not access the same file at the same time.
In practice when you know the Xarray dataset path(s) relevant to your workflow, you may also go directly to the datasets file.
import xarray
extracted_file = "build/tutorial_08_data_extraction/parameter_set0/rectangle_compression_datasets.h5"
group_path = "/RECTANGLE/FieldOutputs/ALL_ELEMENTS"
field_outputs = xarray.open_dataset(extracted_file, group=group_path)
Workflow Visualization#
View the workflow directed graph by running the following command and opening the image in your preferred image viewer. First, plot the workflow with all parameter sets.
$ pwd
/home/roppenheimer/waves-tutorials
$ waves visualize tutorial_08_data_extraction --output-file tutorial_08_data_extraction.png --width=48 --height=12 --exclude-list /usr/bin .stdout .jnl .prt .com .msg .dat .sta
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > waves visualize tutorial_08_data_extraction --output-file tutorial_08_data_extraction.png --width=48 --height=12 --exclude-list .stdout .jnl .prt .com .msg .dat .sta
The output should look similar to the figure below.
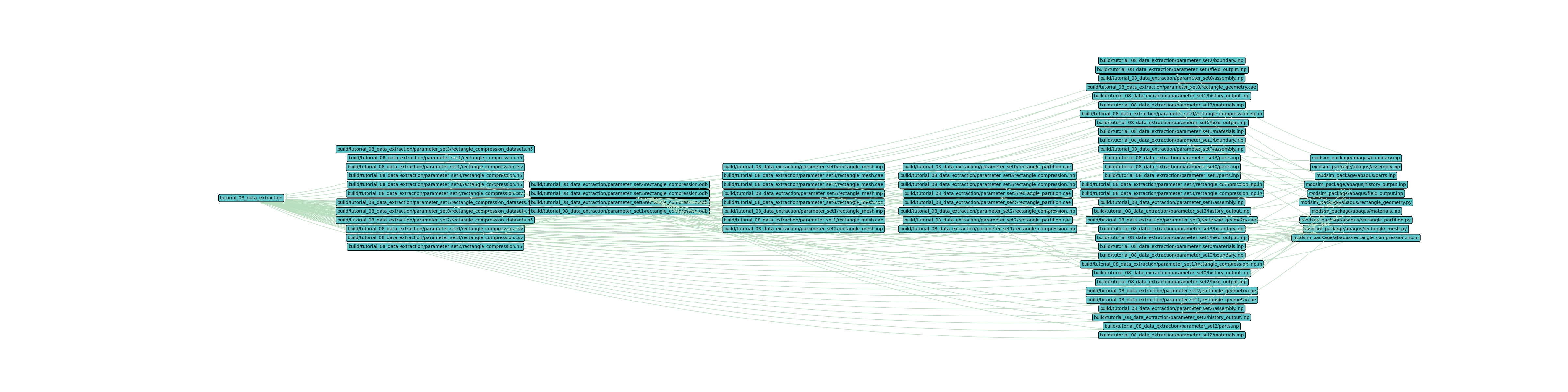
Now plot the workflow with only the first set, set0.
$ pwd
/home/roppenheimer/waves-tutorials
$ waves visualize tutorial_08_data_extraction --output-file tutorial_08_data_extraction_set0.png --width=46 --height=6 --exclude-list /usr/bin .stdout .jnl .prt .com .msg .dat .sta --exclude-regex "set[1-9]"
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > waves visualize tutorial_08_data_extraction --output-file tutorial_08_data_extraction_set0.png --width=46 --height=6 --exclude-list /usr/bin .stdout .jnl .prt .com .msg .dat .sta --exclude-regex "set[1-9]"
The output should look similar to the figure below.
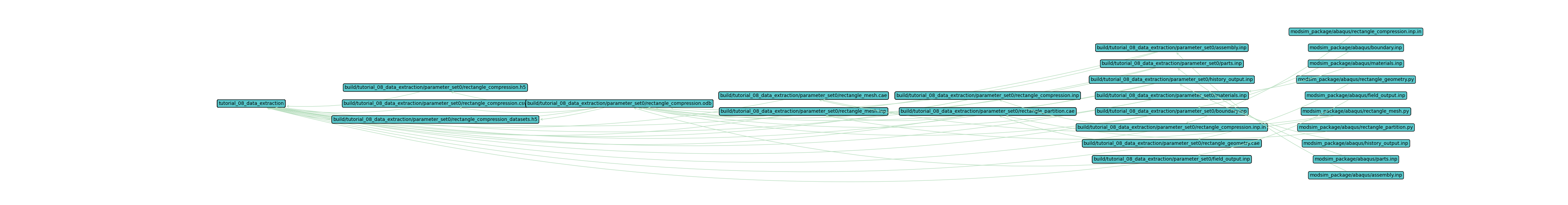
This single set directed graph image should look very similar to the Tutorial 07: Cartesian Product directed
graph. The data extraction step has added the *.h5 files which take the proprietary *.odb data format and
provide h5py and xarray dataset files.
