Tutorial 11: Regression Testing#
Regression testing is the practice of running a verification test suite after making changes to a repository or codebase. For modsim repositories, there may not be many unit or integration tests if there is no software or scripting library specific to the project. Instead, regression testing a modsim repository may look more like regular execution of system tests that verify the simulation workflow still executes as expected.
Ideally, this verification suite of system tests would perform the complete simulation workflow from start to finish. However, modsim repositories often contain simulations that are computationally expensive or produce large amounts of data on disk. In these cases, it may be too expensive to run the full simulation suite at any regular interval. It is still desirable to provide early warning of breaking changes in the simulation workflow, so as much of the workflow that can be tested should be tested as regularly as possible given compute resource constraints.
This tutorial introduces a project wide alias to allow convenient execution of the simulation workflow through the
simulation datacheck task introduced in Tutorial 04: Simulation. From that tutorial onward, each tutorial has
propagated a tutorial specific datacheck alias. This tutorial will add a project wide datacheck alias and apply it
to a copy of the Tutorial 09: Post-Processing configuration files. The user may also go back to previous
tutorials to include the full suite of datacheck tasks in the project wide datacheck regression test alias.
In addition to the datachecks, this tutorial will introduce a full simulation results regression test script and task. The regression test task will be added to the regular workflow alias to run everytime the full workflow is run. This test compares the actual simulation results within a float tolerance. Comprehensive results regression testing is valuable to evaluate changes to important quantities of interest when software versions change, e.g. when installing a new version of Abaqus.
After this tutorial, the workflow will have three sets of tests: fast running unit tests introduced in
Tutorial 10: Unit Testing, relatively fast running simulation preparation checks with the datacheck alias, and
full simulation results regression tests. The tutorial simulations run fast enough that performing the full suite of
tests for every change in the project is tractable. However, in practice, projects may choose to run only the unit tests
and datachecks on a per-change basis and reserve the comprehensive results testing for a scheduled regression suite.
References#
Continuous Integration software
GitHub Actions: https://docs.github.com/en/actions [57]
Gitlab CI: https://docs.gitlab.com/ee/ci/ [58]
Bitbucket Pipelines: https://bitbucket.org/product/features/pipelines [59]
Environment#
SCons and WAVES can be installed in a Conda environment with the Conda package manager. See the Conda installation and Conda environment management documentation for more details about using Conda.
Note
The SALib and numpy versions may not need to be this strict for most tutorials. However, Tutorial: Sensitivity Study uncovered some undocumented SALib version sensitivity to numpy surrounding the numpy v2 rollout.
Create the tutorials environment if it doesn’t exist
$ conda create --name waves-tutorial-env --channel conda-forge waves 'scons>=4.6' matplotlib pandas pyyaml xarray seaborn 'numpy>=2' 'salib>=1.5.1' pytest
PS > conda create --name waves-tutorial-env --channel conda-forge waves scons matplotlib pandas pyyaml xarray seaborn numpy salib pytest
Activate the environment
$ conda activate waves-tutorial-env
PS > conda activate waves-tutorial-env
Some tutorials require additional third-party software that is not available for the Conda package manager. This
software must be installed separately and either made available to SConstruct by modifying your system’s PATH or by
modifying the SConstruct search paths provided to the waves.scons_extensions.add_program() method.
Warning
STOP! Before continuing, check that the documentation version matches your installed package version.
You can find the documentation version in the upper-left corner of the webpage.
You can find the installed WAVES version with
waves --version.
If they don’t match, you can launch identically matched documentation with the WAVES Command-Line Utility
docs subcommand as waves docs.
Directory Structure#
Create and change to a new project root directory to house the tutorial files if you have not already done so. For example
$ mkdir -p ~/waves-tutorials
$ cd ~/waves-tutorials
$ pwd
/home/roppenheimer/waves-tutorials
PS > New-Item $HOME\waves-tutorials -ItemType "Directory"
PS > Set-Location $HOME\waves-tutorials
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
Note
If you skipped any of the previous tutorials, run the following commands to create a copy of the necessary tutorial files.
$ pwd
/home/roppenheimer/waves-tutorials
$ waves fetch --overwrite --tutorial 10 && mv tutorial_10_unit_testing_SConstruct SConstruct
WAVES fetch
Destination directory: '/home/roppenheimer/waves-tutorials'
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > waves fetch --overwrite --tutorial 10 && Move-Item tutorial_10_unit_testing_SConstruct SConstruct -Force
WAVES fetch
Destination directory: 'C:\Users\roppenheimer\waves-tutorials'
Download and copy the
tutorial_09_post_processing.sconsfile to a new file namedtutorial_11_regression_testing.sconswith the WAVES Command-Line Utility fetch subcommand.
$ pwd
/home/roppenheimer/waves-tutorials
$ waves fetch --overwrite tutorials/tutorial_09_post_processing.scons && cp tutorial_09_post_processing.scons tutorial_11_regression_testing.scons
WAVES fetch
Destination directory: '/home/roppenheimer/waves-tutorials'
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > waves fetch --overwrite tutorials\tutorial_09_post_processing.scons && Copy-Item tutorial_09_post_processing.scons tutorial_11_regression_testing.scons
WAVES fetch
Destination directory: 'C:\Users\roppenheimer\waves-tutorials'
Regression Script#
In the
waves-tutorials/modsim_package/pythondirectory, review the file namedregression.pyintroduced in Tutorial 10: Unit Testing.
waves-tutorials/modsim_package/python/regression.py
1#!/usr/bin/env python
2"""Perform regression testing on simulation output."""
3
4import argparse
5import pathlib
6import sys
7
8import pandas
9import yaml
10
11
12def sort_dataframe(
13 dataframe: pandas.DataFrame,
14 index_column: str = "time",
15 sort_columns: list[str] | tuple[str, ...] = ("time", "set_name"),
16) -> pandas.DataFrame:
17 """Return a sorted dataframe and set an index.
18
19 1. sort columns by column name
20 2. sort rows by column values ``sort_columns``
21 3. set an index
22
23 :param dataframe: dataframe to sort
24 :param index_column: name of the column to use an index
25 :param sort_columns: name of the column(s) to sort by
26
27 :returns: sorted and indexed dataframe
28 """
29 return dataframe.reindex(sorted(dataframe.columns), axis=1).sort_values(list(sort_columns)).set_index(index_column)
30
31
32def csv_files_match(
33 current_csv: pandas.DataFrame,
34 expected_csv: pandas.DataFrame,
35 index_column: str = "time",
36 sort_columns: list[str] | tuple[str, ...] = ("time", "set_name"),
37) -> bool:
38 """Compare two pandas DataFrame objects and determine if they match.
39
40 :param current_csv: Current CSV data of generated plot.
41 :param expected_csv: Expected CSV data.
42 :param index_column: name of the column to use an index
43 :param sort_columns: name of the column(s) to sort by. Defaults to ``["time", "set_name"]``
44
45 :returns: True if the CSV files match, False otherwise.
46 """
47 current = sort_dataframe(current_csv, index_column=index_column, sort_columns=sort_columns)
48 expected = sort_dataframe(expected_csv, index_column=index_column, sort_columns=sort_columns)
49 try:
50 pandas.testing.assert_frame_equal(current, expected)
51 except AssertionError as err:
52 print(
53 f"The CSV regression test failed. Data in expected CSV file and current CSV file do not match.\n{err}",
54 file=sys.stderr,
55 )
56 equal = False
57 else:
58 equal = True
59 return equal
60
61
62def main(
63 first_file: pathlib.Path,
64 second_file: pathlib.Path,
65 output_file: pathlib.Path,
66) -> None:
67 """Compare CSV files and return an error code if they differ.
68
69 :param first_file: path-like or file-like object containing the first CSV dataset
70 :param second_file: path-like or file-like object containing the second CSV dataset
71 """
72 regression_results = {}
73
74 # CSV regression file comparison
75 first_data = pandas.read_csv(first_file)
76 second_data = pandas.read_csv(second_file)
77 regression_results.update({"CSV comparison": csv_files_match(first_data, second_data)})
78
79 with output_file.open(mode="w") as output:
80 output.write(yaml.safe_dump(regression_results))
81
82 if len(regression_results.values()) < 1 or not all(regression_results.values()):
83 sys.exit("One or more regression tests failed")
84
85
86def get_parser() -> argparse.ArgumentParser:
87 """Return parser for CLI options.
88
89 All options should use the double-hyphen ``--option VALUE`` syntax to avoid clashes with the Abaqus option syntax,
90 including flag style arguments ``--flag``. Single hyphen ``-f`` flag syntax often clashes with the Abaqus command
91 line options and should be avoided.
92
93 :returns: parser
94 :rtype:
95 """
96 script_name = pathlib.Path(__file__)
97 default_output_file = f"{script_name.stem}.yaml"
98
99 prog = f"python {script_name.name} "
100 cli_description = "Compare CSV files and return an error code if they differ"
101 parser = argparse.ArgumentParser(description=cli_description, prog=prog)
102 parser.add_argument(
103 "FIRST_FILE",
104 type=pathlib.Path,
105 help="First CSV file for comparison",
106 )
107 parser.add_argument(
108 "SECOND_FILE",
109 type=pathlib.Path,
110 help="Second CSV file for comparison",
111 )
112 parser.add_argument(
113 "--output-file",
114 type=pathlib.Path,
115 default=default_output_file,
116 help="Regression test pass/fail list",
117 )
118 return parser
119
120
121if __name__ == "__main__":
122 parser = get_parser()
123 args = parser.parse_args()
124 main(
125 args.FIRST_FILE,
126 args.SECOND_FILE,
127 args.output_file,
128 )
CSV file#
In the
waves-tutorials/modsim_package/pythondirectory, create a new file namedrectangle_compression_cartesian_product.csvfrom the contents below
waves-tutorials/modsim_package/python/rectangle_compression_cartesian_product.csv
1time,set_name,step,elements,integrationPoint,E values,E,S values,S,width,height,global_seed,displacement,set_hash
20.0175000000745058,parameter_set0,Step-1,1,1.0,E22,-0.000159091,S22,-0.0159091,1.1,1.1,1.0,-0.01,181cb00b478ced26819416aeef6a1a3f
30.0175000000745058,parameter_set1,Step-1,1,1.0,E22,-0.000159091,S22,-0.0159091,1.0,1.1,1.0,-0.01,53980146e2729b8956ecf6ef39f342b4
40.0175000000745058,parameter_set2,Step-1,1,1.0,E22,-0.000175,S22,-0.0175,1.0,1.0,1.0,-0.01,cf0934b22f43400165bd3d34aa61013f
50.0175000000745058,parameter_set3,Step-1,1,1.0,E22,-0.000175,S22,-0.0175,1.1,1.0,1.0,-0.01,fff52e9be95e50cc872ec321280d91a7
60.0709374994039536,parameter_set0,Step-1,1,1.0,E22,-0.000644886,S22,-0.0644886,1.1,1.1,1.0,-0.01,181cb00b478ced26819416aeef6a1a3f
70.0709374994039536,parameter_set1,Step-1,1,1.0,E22,-0.000644886,S22,-0.0644886,1.0,1.1,1.0,-0.01,53980146e2729b8956ecf6ef39f342b4
80.0709374994039536,parameter_set2,Step-1,1,1.0,E22,-0.000709375,S22,-0.0709375,1.0,1.0,1.0,-0.01,cf0934b22f43400165bd3d34aa61013f
90.0709374994039536,parameter_set3,Step-1,1,1.0,E22,-0.000709375,S22,-0.0709375,1.1,1.0,1.0,-0.01,fff52e9be95e50cc872ec321280d91a7
100.251289069652557,parameter_set0,Step-1,1,1.0,E22,-0.00228445,S22,-0.228445,1.1,1.1,1.0,-0.01,181cb00b478ced26819416aeef6a1a3f
110.251289069652557,parameter_set1,Step-1,1,1.0,E22,-0.00228445,S22,-0.228445,1.0,1.1,1.0,-0.01,53980146e2729b8956ecf6ef39f342b4
120.251289069652557,parameter_set2,Step-1,1,1.0,E22,-0.00251289,S22,-0.251289,1.0,1.0,1.0,-0.01,cf0934b22f43400165bd3d34aa61013f
130.251289069652557,parameter_set3,Step-1,1,1.0,E22,-0.00251289,S22,-0.251289,1.1,1.0,1.0,-0.01,fff52e9be95e50cc872ec321280d91a7
140.859975576400757,parameter_set0,Step-1,1,1.0,E22,-0.00781796,S22,-0.781796,1.1,1.1,1.0,-0.01,181cb00b478ced26819416aeef6a1a3f
150.859975576400757,parameter_set1,Step-1,1,1.0,E22,-0.00781796,S22,-0.781796,1.0,1.1,1.0,-0.01,53980146e2729b8956ecf6ef39f342b4
160.859975576400757,parameter_set2,Step-1,1,1.0,E22,-0.00859976,S22,-0.859976,1.0,1.0,1.0,-0.01,cf0934b22f43400165bd3d34aa61013f
170.859975576400757,parameter_set3,Step-1,1,1.0,E22,-0.00859976,S22,-0.859976,1.1,1.0,1.0,-0.01,fff52e9be95e50cc872ec321280d91a7
181.0,parameter_set0,Step-1,1,1.0,E22,-0.00909091,S22,-0.909091,1.1,1.1,1.0,-0.01,181cb00b478ced26819416aeef6a1a3f
191.0,parameter_set1,Step-1,1,1.0,E22,-0.00909091,S22,-0.909091,1.0,1.1,1.0,-0.01,53980146e2729b8956ecf6ef39f342b4
201.0,parameter_set2,Step-1,1,1.0,E22,-0.01,S22,-1.0,1.0,1.0,1.0,-0.01,cf0934b22f43400165bd3d34aa61013f
211.0,parameter_set3,Step-1,1,1.0,E22,-0.01,S22,-1.0,1.1,1.0,1.0,-0.01,fff52e9be95e50cc872ec321280d91a7
This file represents a copy of previous simulation results that the project has stored as the reviewed and approved
simulation results. The regression task will compare these “past” results with the current simulation results produced
during the post-processing task introduced in Tutorial 09: Post-Processing
rectangle_compression_cartesian_product.csv using the regression.py CLI.
SConscript#
A diff against the tutorial_09_post_processing.scons file from Tutorial 09: Post-Processing is included below to help identify the
changes made in this tutorial.
waves-tutorials/tutorial_11_regression_testing.scons
--- /home/runner/work/waves/waves/build/docs/tutorials_tutorial_09_post_processing.scons
+++ /home/runner/work/waves/waves/build/docs/tutorials_tutorial_11_regression_testing.scons
@@ -5,6 +5,8 @@
* ``env`` - The SCons construction environment with the following required keys
+ * ``datacheck_alias`` - String for the alias collecting the datacheck workflow targets
+ * ``regression_alias`` - String for the alias collecting the regression test suite targets
* ``unconditional_build`` - Boolean flag to force building of conditionally ignored targets
* ``abaqus`` - String path for the Abaqus executable
"""
@@ -176,11 +178,27 @@
)
)
+# Regression test
+workflow.extend(
+ env.PythonScript(
+ target=["regression.yaml"],
+ source=[
+ "#/modsim_package/python/regression.py",
+ "stress_strain_comparison.csv",
+ "#/modsim_package/python/rectangle_compression_cartesian_product.csv",
+ ],
+ subcommand_options="${SOURCES[1:].abspath} --output-file ${TARGET.abspath}",
+ )
+)
+
# Collector alias based on parent directory name
env.Alias(workflow_name, workflow)
env.Alias(f"{workflow_name}_datacheck", datacheck)
+env.Alias(env["datacheck_alias"], datacheck)
+env.Alias(env["regression_alias"], datacheck)
if not env["unconditional_build"] and not env["ABAQUS_PROGRAM"]:
print(f"Program 'abaqus' was not found in construction environment. Ignoring '{workflow_name}' target(s)")
Ignore([".", workflow_name], workflow)
Ignore([".", f"{workflow_name}_datacheck"], datacheck)
+ Ignore([".", env["datacheck_alias"], env["regression_alias"]], datacheck)
There are two changes made in this tutorial. The first is to compare the expected simulation results to the current simulation’s output. A new task compares the expected results as the CSV file created above against the current simulation output with the new Python script, regression.py. See the regression.py CLI documentation for a description of the post-processing script’s behavior.
The second change adds a dedicated alias for the datacheck targets to allow partial workflow execution. This is useful
when a full simulation may take a long time, but the simulation preparation is worth testing on a regular basis. We’ve
also added the regression alias introduced briefly in Tutorial 10: Unit Testing. Previously, this alias was a
duplicate of the unit_testing workflow alias. Now this alias can be used as a collector alias for running a
regression suite with a single command, while preserving the ability to run the unit tests as a standalone workflow.
Here we add the datacheck targets as an example of running a partial workflow as part of the regression test suite. For
fast running simulations, it would be valuable to run the full simulation and post-processing with CSV results testing
as part of the regular regression suite. For large projects with long running simulations, several regression aliases
may be added to facilitate testing at different intervals. For instance, the datachecks might be run everytime the
project changes, but the simulations might be run on a weekly schedule with a regression_weekly alias that includes
the full simulations in addition to the unit tests and datachecks.
SConstruct#
A diff against the SConstruct file from Tutorial 10: Unit Testing is included below to help identify the
changes made in this tutorial.
waves-tutorials/SConstruct
--- /home/runner/work/waves/waves/build/docs/tutorials_tutorial_10_unit_testing_SConstruct
+++ /home/runner/work/waves/waves/build/docs/tutorials_tutorial_11_regression_testing_SConstruct
@@ -1,5 +1,5 @@
#! /usr/bin/env python
-"""Configure the WAVES unit testing tutorial."""
+"""Configure the WAVES regression testing tutorial."""
import os
import pathlib
@@ -83,6 +83,7 @@
"project_dir": project_dir,
"version": version,
"regression_alias": "regression",
+ "datacheck_alias": "datacheck",
}
for key, value in project_variables.items():
env[key] = value
@@ -113,6 +114,7 @@
"tutorial_07_cartesian_product.scons",
"tutorial_08_data_extraction.scons",
"tutorial_09_post_processing.scons",
+ "tutorial_11_regression_testing.scons",
]
for workflow in workflow_configurations:
build_dir = env["variant_dir_base"] / pathlib.Path(workflow).stem
Build Targets#
Build the datacheck targets without executing the full simulation workflow
$ pwd
/home/roppenheimer/waves-tutorials
$ time scons datacheck --jobs=4
<output truncated>
scons: done building targets.
real 0m9.952s
user 0m21.537s
sys 0m15.664s
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > Measure-Command { scons datacheck --jobs=4 | Out-Default }
<output truncated>
scons: done building targets.
Days : 0
Hours : 0
Minutes : 0
Seconds : 10
Milliseconds : 129
Ticks : 101291696
TotalDays : 0.0001172357592593
TotalHours : 0.002813658222222
TotalMinutes : 0.1688194933333
TotalSeconds : 10.1291696
TotalMilliseconds : 10129.1696
Run the full workflow and verify that the CSV regression test passes
$ pwd
/home/roppenheimer/waves-tutorials
$ scons datacheck --clean
$ time scons tutorial_11_regression_testing --jobs=4
<output truncated>
scons: done building targets.
real 0m29.031s
user 0m25.712s
sys 0m25.622s
$ cat build/tutorial_11_regression_testing/regression.yaml
CSV comparison: true
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > scons datacheck --clean
PS > Measure-Command { scons tutorial_11_regression_testing --jobs=4 | Out-Default }
<output truncated>
scons: done building targets.
Days : 0
Hours : 0
Minutes : 0
Seconds : 31
Milliseconds : 493
Ticks : 314936692
TotalDays : 0.0003645100601852
TotalHours : 0.008748241444444
TotalMinutes : 0.5248944866667
TotalSeconds : 31.4936692
TotalMilliseconds : 31493.6692
PS > Get-Content build\tutorial_11_regression_testing\regression.yaml
CSV comparison: true
If you haven’t added the project-wide datacheck alias to the previous tutorials, you should expect the datacheck
alias to run faster than the tutorial_11_regression_testing alias because the datacheck excludes the solve, extract,
and post-processing tasks. In these tutorials, the difference in execution time is not large. However, in many
production modsim projects, the simulations may require hours or even days to complete. In that case, the relatively
fast running solverprep verification may be tractable for regular testing where the full simulations and post-processing
are not.
To approximate the time savings of the new project-wide datacheck alias for a (slightly) larger modsim project, you
can go back through the previous tutorials and add each tutorial’s datacheck task to the new alias. For a fair
comparison, you will also need to add a comparable alias to collect the full workflow for each tutorial, e.g.
full_workflows. You can then repeat the time commands above with a more comprehensive datacheck and
full_workflows aliases.
Output Files#
$ pwd
/home/roppenheimer/waves-tutorials
$ tree build/tutorial_11_regression_testing/parameter_set0/
build/tutorial_11_regression_testing/parameter_set0/
|-- abaqus.rpy
|-- abaqus.rpy.1
|-- abaqus.rpy.2
|-- assembly.inp
|-- boundary.inp
|-- field_output.inp
|-- history_output.inp
|-- materials.inp
|-- parts.inp
|-- rectangle_compression.inp
|-- rectangle_compression.inp.in
|-- rectangle_compression_DATACHECK.023
|-- rectangle_compression_DATACHECK.com
|-- rectangle_compression_DATACHECK.dat
|-- rectangle_compression_DATACHECK.mdl
|-- rectangle_compression_DATACHECK.msg
|-- rectangle_compression_DATACHECK.odb
|-- rectangle_compression_DATACHECK.prt
|-- rectangle_compression_DATACHECK.sim
|-- rectangle_compression_DATACHECK.stdout
|-- rectangle_compression_DATACHECK.stt
|-- rectangle_geometry.cae
|-- rectangle_geometry.jnl
|-- rectangle_geometry.stdout
|-- rectangle_mesh.cae
|-- rectangle_mesh.inp
|-- rectangle_mesh.jnl
|-- rectangle_mesh.stdout
|-- rectangle_partition.cae
|-- rectangle_partition.jnl
`-- rectangle_partition.stdout
0 directories, 31 files
$ tree build/tutorial_11_regression_testing/ -L 1
build/tutorial_11_regression_testing/
|-- parameter_set0
|-- parameter_set1
|-- parameter_set2
|-- parameter_set3
|-- parameter_study.h5
|-- regression.yaml
|-- regression.yaml.stdout
|-- stress_strain_comparison.csv
|-- stress_strain_comparison.pdf
`-- stress_strain_comparison.pdf.stdout
4 directories, 6 files
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > tree build\tutorial_11_regression_testing\parameter_set0\ /F
C:\USERS\ROPPENHEIMER\WAVES-TUTORIALS\BUILD\TUTORIAL_11_REGRESSION_TESTING\PARAMETER_SET0
abaqus.rpy
abaqus.rpy.2
abaqus.rpy.3
abaqus.rpy.4
abaqus.rpy.5
assembly.inp
boundary.inp
field_output.inp
history_output.inp
materials.inp
parts.inp
rectangle_compression.com
rectangle_compression.csv
rectangle_compression.dat
rectangle_compression.h5
rectangle_compression.inp
rectangle_compression.inp.in
rectangle_compression.msg
rectangle_compression.odb
rectangle_compression.odb.stdout
rectangle_compression.prt
rectangle_compression.sta
rectangle_compression_DATACHECK.cax
rectangle_compression_datasets.h5
rectangle_geometry.cae
rectangle_geometry.cae.stdout
rectangle_geometry.jnl
rectangle_mesh.cae
rectangle_mesh.inp
rectangle_mesh.inp.stdout
rectangle_mesh.jnl
rectangle_partition.cae
rectangle_partition.cae.stdout
rectangle_partition.jnl
No subfolders exist
PS > Get-ChildItem build\tutorial_11_regression_testing\
Directory: C:\Users\roppenheimer\waves-tutorials\build\tutorial_11_regression_testing
Mode LastWriteTime Length Name
---- ------------- ------ ----
d---- 6/9/2023 4:32 PM parameter_set0
d---- 6/9/2023 4:32 PM parameter_set1
d---- 6/9/2023 4:32 PM parameter_set2
d---- 6/9/2023 4:32 PM parameter_set3
-a--- 6/9/2023 4:32 PM 9942 parameter_study.h5
-a--- 6/9/2023 4:32 PM 22 regression.yaml
-a--- 6/9/2023 4:32 PM 0 regression.yaml.stdout
-a--- 6/9/2023 4:32 PM 2609 stress_strain_comparison.csv
-a--- 6/9/2023 4:32 PM 12061 stress_strain_comparison.pdf
-a--- 6/9/2023 4:32 PM 1160 stress_strain_comparison.pdf.stdout
Workflow Visualization#
View the workflow directed graph by running the following command and opening the image in your preferred image viewer.
Plot the workflow with only the first set, set0.
$ pwd
/home/roppenheimer/waves-tutorials
$ waves visualize datacheck --output-file tutorial_11_datacheck_set0.png --width=42 --height=8 --exclude-list /usr/bin .stdout .jnl .prt .com .msg .dat .sta --exclude-regex "set[1-9]"
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
PS > waves visualize datacheck --output-file tutorial_11_datacheck_set0.png --width=42 --height=8 --exclude-list .stdout .jnl .prt .com .msg .dat .sta --exclude-regex "set[1-9]"
The output should look similar to the figure below.
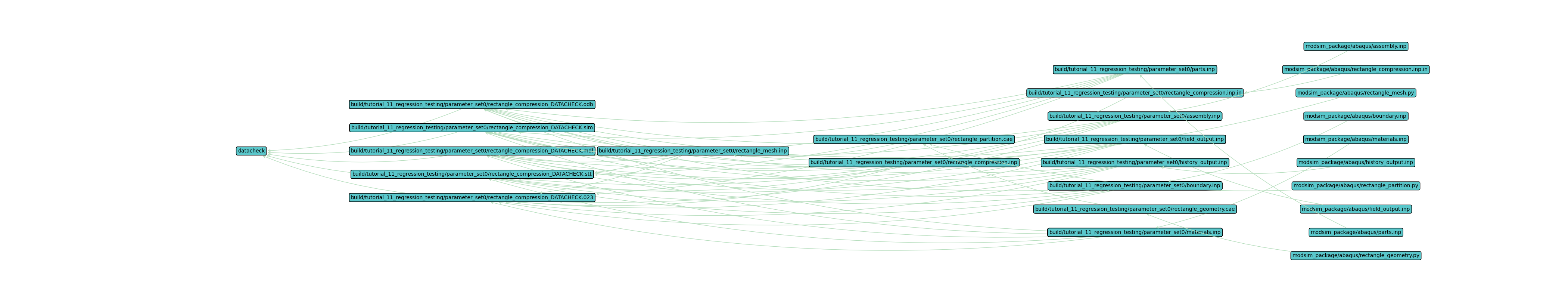
This tutorial’s datacheck directed graph should look different from the graph in
Tutorial 09: Post-Processing. Here we have plotted the datacheck alias output, which does not execute the
full simulation workflow. This partial directed graph may run faster than the full simulation workflow for frequent
regression tests.
Automation#
There are many tools that can help automate the execution of the modsim project regression tests. With the collector
alias, those tools need only execute a single SCons command to perform the selected, lower cost tasks for simulation
workflow verification, scons datacheck. If git [21] is used as the version control system, developer
operations software such as GitHub [24], Gitlab [25], and Atlassian’s Bitbucket
[26] provide continuous integration software that can automate verification tests on triggers, such as
merge requests, or on a regular schedule.
