Quickstart: Gmsh+CalculiX#
This tutorial mirrors the Abaqus-based Quickstart: Abaqus with a fully open-source, conda-forge workflow. The geometry is created and meshed with Gmsh. The finite element simulation is performed with CalculiX. Finally, the data is extracted with ccx2paraview and meshio for scripted quantity of interest (QoI) calculations and post-processing.
This quickstart will create a minimal, two file project configuration combining elements of the tutorials listed below.
These tutorials and this quickstart describe the computational engineering workflow through simulation execution and post-processing. This tutorial will use a different working directory and directory structure than the rest of the tutorials to avoid filename clashes. The quickstart also uses a flat directory structure to simplify the project configuration. Larger projects, like the ModSim Templates, may require a hierarchical directory structure to separate files with identical basenames.
References#
Environment#
Warning
The Gmsh+CalculiX tutorial requires a different compute environment than the other tutorials. The following commands
create a dedicated environment for the use of this tutorial. You can also use your existing tutorial environment with
the conda install command while your environment is active.
All of the software required for this tutorial can be installed in a Conda environment with the Conda package manager. See the Conda installation and Conda environment management documentation for more details about using Conda.
Create the Gmsh+CalculiX tutorial environment if it doesn’t exist
$ conda create --name waves-gmsh-env --channel conda-forge waves 'scons>=4.6' python-gmsh calculix 'ccx2paraview>=3.2' meshio
PS > conda create --name waves-gmsh- --channel conda-forge waves scons python-gmsh calculix ccx2paraview meshio
Activate the environment
$ conda activate waves-gmsh-env
PS > conda activate waves-gmsh-env
Warning
STOP! Before continuing, check that the documentation version matches your installed package version.
You can find the documentation version in the upper-left corner of the webpage.
You can find the installed WAVES version with
waves --version.
If they don’t match, you can launch identically matched documentation with the WAVES Command-Line Utility
docs subcommand as waves docs.
Directory Structure#
Create and change to a new project root directory to house the tutorial files if you have not already done so. For example
$ mkdir -p ~/waves-tutorials
$ cd ~/waves-tutorials
$ pwd
/home/roppenheimer/waves-tutorials
PS > New-Item $HOME\waves-tutorials -ItemType "Directory"
PS > Set-Location $HOME\waves-tutorials
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials
Create a new
tutorial_gmshdirectory with thewaves fetchcommand below
$ waves fetch tutorials/tutorial_gmsh --destination ~/waves-tutorials/tutorial_gmsh
WAVES fetch
Destination directory: '/home/roppenheimer/waves-tutorials/tutorial_gmsh'
$ cd ~/waves-tutorials/tutorial_gmsh
$ pwd
/home/roppenheimer/waves-tutorials/tutorial_gmsh
$ ls .
environment.yml post_processing.py rectangle_compression.inp.in rectangle.py SConscript SConstruct strip_heading.py time_points.inp vtu2xarray.py
PS > waves fetch tutorials\tutorial_gmsh --destination $HOME\waves-tutorials\tutorial_gmsh
WAVES fetch
Destination directory: 'C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh'
PS > Set-Location $HOME\waves-tutorials\tutorial_gmsh
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh
PS > Get-ChildItem .
Directory: C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 329 environment.yml
-a--- 6/9/2023 4:32 PM 9250 post_processing.py
-a--- 6/9/2023 4:32 PM 498 rectangle_compression.inp.in
-a--- 6/9/2023 4:32 PM 5818 rectangle.py
-a--- 6/9/2023 4:32 PM 2280 SConscript
-a--- 6/9/2023 4:32 PM 3073 SConstruct
-a--- 6/9/2023 4:32 PM 1206 strip_heading.py
-a--- 6/9/2023 4:32 PM 44 time_points.inp
-a--- 6/9/2023 4:32 PM 6079 vtu2xarray.py
SConscript#
Managing digital data and workflows in modern computational science and engineering is a difficult and error-prone task. The large number of steps in the workflow and complex web of interactions between data files results in non-intuitive dependencies that are difficult to manage by hand. This complexity grows substantially when the workflow includes parameter studies. WAVES enables the use of traditional software build systems in computational science and engineering workflows with support for common engineering software and parameter study management.
Build systems construct a directed acyclic graph (DAG) from small, individual task definitions. Each task is defined by the developer and subsequently linked by the build system. Tasks are composed of targets, sources, and actions. A target is the output of the task. Sources are the required direct-dependency files used by the task and may be files tracked by the version control system for the project or files produced by other tasks. Actions are the executable commands that produce the target files. In pseudocode, this might look like a dictionary:
task1:
target: output1
source: source1
action: action1 --input source1 --output output1
task2:
target: output2
source: output1
action: action2 --input output1 --output output2
As the number of discrete tasks increases, and as cross-dependencies grow, an automated tool to construct the build order becomes more important. Besides simplifying the process of constructing the workflow DAG, most build systems also incorporate a state machine. The build system tracks the execution state of the DAG and will only re-build out-of-date portions of the DAG. This is especially valuable when trouble-shooting or expanding a workflow. For instance, when adding or modifying the post-processing step, the build system will not re-run simulation tasks that are computationally expensive and require significant wall time to solve.
The SConscript file below contains the workflow task definitions. Review the source and target files defining the
workflow tasks. As discussed briefly above and in detail in Build System, a task definition also requires an
action. For convenience, WAVES provides builders for common engineering software with pre-defined task actions. See
the waves.scons_extensions.abaqus_journal_builder_factory() and
waves.scons_extensions.abaqus_solver_builder_factory() for more complete descriptions of the builder actions.
waves-tutorials/tutorial_gmsh/SConscript
1#! /usr/bin/env python
2"""Configure the WAVES Gmsh tutorial workflow."""
3
4import vtu2xarray
5
6Import("env", "alias", "parameters")
7
8# Geometry, Partition, Mesh
9env.PythonScript(
10 target=["rectangle_gmsh.inp"],
11 source=["rectangle.py"],
12 subcommand_options="--output-file=${TARGET.abspath}",
13 **parameters,
14)
15
16env.PythonScript(
17 target=["rectangle_mesh.inp"],
18 source=["strip_heading.py", "rectangle_gmsh.inp"],
19 subcommand_options="--input-file=${SOURCES[1].abspath} --output-file=${TARGET.abspath}",
20)
21
22# SolverPrep
23env.CopySubstfile(
24 ["#/rectangle_compression.inp.in"],
25 substitution_dictionary=env.SubstitutionSyntax(parameters),
26)
27
28# CalculiX Solve
29env.CalculiX(
30 target=[f"rectangle_compression.{suffix}" for suffix in ("frd", "dat", "sta", "cvg", "12d")],
31 source=["rectangle_compression.inp"],
32)
33
34# Extract
35time_points_source_file = env["project_directory"] / "time_points.inp"
36time_points = vtu2xarray.time_points_from_file(time_points_source_file)
37time_point_indices = range(1, len(time_points) + 1, 1)
38vtu_files = [f"rectangle_compression.{increment:02}.vtu" for increment in time_point_indices]
39env.Command(
40 target=[*vtu_files, "rectangle_compression.vtu.stdout"],
41 source=["rectangle_compression.frd"],
42 action=["cd ${TARGET.dir.abspath} && ccx2paraview ${SOURCES[0].abspath} vtu > ${TARGETS[-1].abspath} 2>&1"],
43)
44env.PythonScript(
45 target=["rectangle_compression.h5"],
46 source=["vtu2xarray.py", "time_points.inp", vtu_files],
47 subcommand_options=(
48 "--input-file ${SOURCES[2:].abspath} --output-file ${TARGET.abspath} --time-points-file ${SOURCES[1].abspath}"
49 ),
50)
51
52
53# Post-processing
54target = env.PythonScript(
55 target=["stress_strain.pdf", "stress_strain.csv"],
56 source=["post_processing.py", "rectangle_compression.h5"],
57 subcommand_options=(
58 "--input-file ${SOURCES[1:].abspath} --output-file ${TARGET.abspath} --x-units mm/mm --y-units MPa"
59 ),
60)
61
62# Collector alias named after the model simulation
63env.Alias(alias, target)
64
65if not env["unconditional_build"] and (not env["CCX_PROGRAM"] or not env["ccx2paraview"]):
66 print(
67 "Program 'CalculiX (ccx)' or 'ccx2paraview' was not found in construction environment. "
68 "Ignoring 'rectangle' target(s)"
69 )
70 Ignore([".", alias], target)
SConstruct#
For this quickstart, we will not discuss the main SCons configuration file, named SConstruct, in detail.
Tutorial 00: SConstruct has a more complete discussion about the contents of the SConstruct file.
One of the primary benefits to WAVES is the ability to robustly integrate the conditional re-building behavior of a build system with computational parameter studies. Because most build systems consist of exactly two steps: configuration and execution, the full DAG must be fixed at configuration time. To avoid hardcoding the parameter study tasks, it is desirable to re-use the existing workflow or task definitions. This could be accomplished with a simple for-loop and naming convention; however, it is common to run a small, scoping parameter study prior to exploring the full parameter space which would require careful set re-numbering to preserve previous work.
To avoid out-of-sync errors in parameter set definitions when updating a previously executed parameter study, WAVES provides a parameter study generator utility that uniquely identifies parameter sets by contents, assigns a unique index to each parameter set, and guarantees that previously executed sets are matched to their unique identifier. When expanding or re-executing a parameter study, WAVES enforces set name/content consistency which in turn ensures that the build system can correctly identify previous work and only re-build the new or changed sets.
In the configuration snippet below, the workflow parameterization is performed in the root configuration file,
SConstruct. This allows us to re-use the entire workflow file, SConscript, with more than one parameter study.
First, we define a nominal workflow. Nominal workflows can be defined as a simple dictionary. This can be useful for
trouble-shooting the workflow, simulation definition, and simulation convergence prior to running a larger parameter
study. Second, we define a small mesh convergence study where the only parameter that changes is the mesh global seed.
waves-tutorials/tutorial_gmsh/SConstruct
61# Define parameter studies
62nominal_parameters = {
63 "width": 1.0,
64 "height": 1.0,
65 "global_seed": 1.0,
66 "displacement": -0.01,
67}
68mesh_convergence_parameter_study_file = env["parameter_study_directory"] / "mesh_convergence.h5"
69mesh_convergence_parameter_generator = waves.parameter_generators.CartesianProduct(
70 {
71 "width": [1.0],
72 "height": [1.0],
73 "global_seed": [1.0, 0.5, 0.25, 0.125],
74 "displacement": [-0.01],
75 },
76 output_file=mesh_convergence_parameter_study_file,
77 previous_parameter_study=mesh_convergence_parameter_study_file,
78)
79mesh_convergence_parameter_generator.write()
Finally, we call the workflow SConscript file in a loop where the study names and definitions are unpacked into the
workflow call. The ParameterStudySConscript method handles the differences between a nominal parameter set
dictionary and the mesh convergence parameter study object. The SConscript file has been written to accept the
parameters variable that will be unpacked by this function.
waves-tutorials/tutorial_gmsh/SConstruct
81# Add workflow(s)
82workflow_configurations = [
83 ("nominal", nominal_parameters),
84 ("mesh_convergence", mesh_convergence_parameter_generator),
85]
86for study_name, study_definition in workflow_configurations:
87 env.ParameterStudySConscript(
88 "SConscript",
89 variant_dir=build_directory / study_name,
90 exports={"env": env, "alias": study_name},
91 study=study_definition,
92 subdirectories=True,
93 duplicate=True,
94 )
In this tutorial, the entire workflow is re-run from scratch for each parameter set. This simplifies the parameter study
construction and enables the geometric parameterization hinted at in the width and height parameters. Not all
workflows require the same level of granularity and re-use. There are resource trade-offs to workflow construction, task
definition granularity, and computational resources. For instance, if the geometry and partition tasks required
significant wall time, but are not part of the mesh convergence study, it might be desirable to parameterize within the
SConscript file where the geometry and partition tasks could be excluded from the parameter study.
WAVES provides several solutions for parameterizing at the level of workflow files, task definitions, or in arbitrary locations and methods, depending on the needs of the project.
workflow files:
waves.scons_extensions.parameter_study_sconscript()task definitions:
waves.scons_extensions.parameter_study_task()anywhere:
waves.parameter_generators.ParameterGenerator.parameter_study_to_dict()
Build Targets#
In SConstruct, the workflows were provided aliases matching the study names for more convenient execution. First,
run the nominal workflow and observe the task command output as below. The default behavior of SCons is to report
each task’s action as it is executed. WAVES builders capture the STDOUT and STDERR into per-task log files to aid in
troubleshooting and to remove clutter from the terminal output. On first execution you may see a warning message as a
previous parameter study is being requested which will only exist on subsequent executions.
$ pwd
/home/roppenheimer/waves-tutorials/tutorial_gmsh
$ scons nominal
scons: Reading SConscript files ...
Checking whether 'ccx' program exists.../projects/aea_compute/waves-env/bin/ccx
Checking whether 'ccx2paraview' program exists.../projects/aea_compute/waves-env/bin/ccx2paraview
scons: done reading SConscript files.
scons: Building targets ...
Copy("build/nominal/rectangle_compression.inp.in", "rectangle_compression.inp.in")
Creating 'build/nominal/rectangle_compression.inp'
cd /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal && python /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle.py --output-file=/home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_gmsh.inp > /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_gmsh.inp.stdout 2>&1
cd /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal && python /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/strip_heading.py --input-file=/home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_gmsh.inp --output-file=/home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_mesh.inp > /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_mesh.inp.stdout 2>&1
cd /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal && /projects/aea_compute/waves-env/bin/ccx -i rectangle_compression > /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.frd.stdout 2>&1
cd /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal && ccx2paraview /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.frd vtu > /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.vtu.stdout 2>&1
cd /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal && python /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/vtu2xarray.py --input-file /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.01.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.02.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.03.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.04.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.05.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.06.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.07.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.08.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.09.vtu /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.10.vtu --output-file /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.h5 --time-points-file /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/time_points.inp > /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.h5.stdout 2>&1
cd /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal && python /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/post_processing.py --input-file /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_compression.h5 --output-file /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/stress_strain_comparison.pdf --x-units mm/mm --y-units MPa > /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/stress_strain_comparison.pdf.stdout 2>&1
scons: done building targets.
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh
PS > scons nominal
scons: Reading SConscript files ...
Checking whether 'ccx' program exists...C:\Users\oppenheimer\AppData\Local\miniconda3\envs\waves-gmsh-env\Library\bin\ccx.EXE
Checking whether 'ccx2paraview' program exists...C:\Users\oppenheimer\AppData\Local\miniconda3\envs\waves-gmsh-env\Scripts\ccx2paraview.EXE
scons: done reading SConscript files.
scons: Building targets ...
Copy("build\nominal\rectangle_compression.inp.in", "rectangle_compression.inp.in")
Creating 'build\nominal\rectangle_compression.inp'
cd C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal && python C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle.py --output-file=C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_gmsh.inp > C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_gmsh.inp.stdout 2>&1
cd C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal && python C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\strip_heading.py --input-file=C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_gmsh.inp --output-file=C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_mesh.inp > C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_mesh.inp.stdout 2>&1
cd C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal && C:\Users\oppenheimer\AppData\Local\miniconda3\envs\waves-gmsh-env\Library\bin\ccx.EXE -i rectangle_compression > C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.frd.stdout 2>&1
cd C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal && ccx2paraview C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.frd vtu > C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.vtu.stdout 2>&1
cd C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal && python C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\vtu2xarray.py --input-file C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.01.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.02.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.03.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.04.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.05.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.06.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.07.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.08.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.09.vtu C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.10.vtu --output-file C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.h5 --time-points-file C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\time_points.inp > C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.h5.stdout 2>&1
cd C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal && python C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\post_processing.py --input-file C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_compression.h5 --output-file C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\stress_strain.pdf --x-units mm/mm --y-units MPa > C:\Users\oppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\stress_strain.pdf.stdout 2>&1
scons: done building targets.
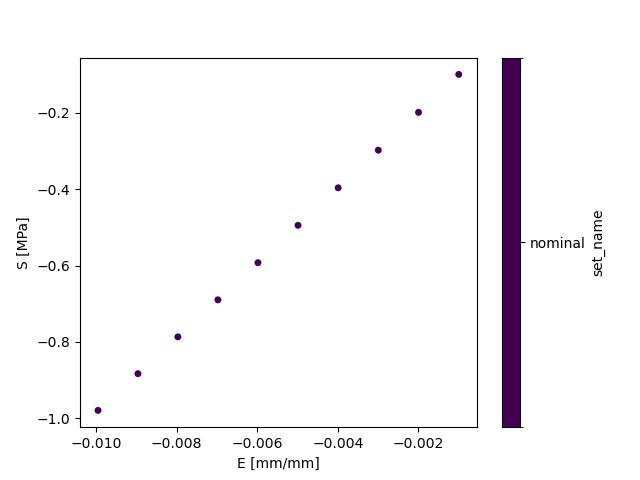
You will find the output files in the build directory. The final post-processing image of the uniaxial compression
stress-strain curve is found in stress_strain.pdf.
$ pwd
/home/roppenheimer/waves-tutorials/tutorial_gmsh
$ ls build/nominal/stress_strain.pdf
build/nominal/stress_strain.pdf
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh
PS > Get-ChildItem build\nominal\stress_strain.pdf
Directory: C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh\build\nominal
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 12951 stress_strain.pdf
Before running the parameter study, explore the conditional re-build behavior of the workflow by deleting the
intermediate output file rectangle_mesh.inp and re-executing the workflow. You should observe that only the command
which produces the orphan mesh rectangle_mesh.inp file is re-run. You can confirm by inspecting the time stamps of
files in the build directory before and after file removal and workflow execution.
$ pwd
/home/roppenheimer/waves-tutorials/tutorial_gmsh
$ rm build/nominal/rectangle_mesh.inp
$ scons nominal
scons: Reading SConscript files ...
Checking whether 'ccx' program exists.../projects/aea_compute/waves-env/bin/ccx
Checking whether 'ccx2paraview' program exists.../projects/aea_compute/waves-env/bin/ccx2paraview
scons: done reading SConscript files.
scons: Building targets ...
cd /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal && python /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/strip_heading.py --input-file=/home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_gmsh.inp --output-file=/home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_mesh.inp > /home/roppenheimer/waves-tutorials/tutorial_gmsh/build/nominal/rectangle_mesh.inp.stdout 2>&1
scons: `nominal' is up to date.
scons: done building targets.
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh
PS > Remove-Item build\nominal\rectangle_mesh.inp
PS > scons nominal
scons: Reading SConscript files ...
Checking whether 'ccx' program exists...C:\Users\roppenheimer\AppData\Local\miniconda3\envs\waves-gmsh-env\Library\bin\ccx.EXE
Checking whether 'ccx2paraview' program exists...C:\Users\roppenheimer\AppData\Local\miniconda3\envs\waves-gmsh-env\Scripts\ccx2paraview.EXE
scons: done reading SConscript files.
scons: Building targets ...
cd C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh\build\nominal && python C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\strip_heading.py --input-file=C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_gmsh.inp --output-file=C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_mesh.inp > C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh\build\nominal\rectangle_mesh.inp.stdout 2>&1
scons: `nominal' is up to date.
scons: done building targets.
You should expect that the geometry and partition tasks do not need to re-execute because their output files still exist and they are upstream of the mesh task. But the tasks after the mesh task did not re-execute, either. This should be somewhat surprising. The simulation itself depends on the mesh file, so why didn’t the workflow re-execute all tasks from mesh to post-processing?
Many software build systems, such as GNU Make use file system modification time stamps to track DAG state
[30]. By default the SCons state machine uses file signatures built from md5 hashes to identify task
state. If the contents of the rectangle_mesh.inp file do not change, then the md5 signature on re-execution still
matches the build state for the rest of the downstream tasks, which do not need to rebuild.
This default behavior of SCons makes it desirable for computational science and engineering workflows where downstream tasks may be computationally expensive. The added cost of computing md5 signatures during configuration is valuable if it prevents re-execution of a computationally expensive simulation. In actual practice, production engineering analysis workflows may include tasks and simulations with wall clock run times of hours to days. By comparison, the cost of using md5 signatures instead of time stamps is often negligible.
Now run the mesh convergence study as below. SCons uses the --jobs option to control the number of threads used
in task execution and will run up to 4 tasks simultaneously.
$ scons mesh_convergence --jobs=4
...
PS > scons mesh_convergence --jobs=4
...
The output is truncated, but should look very similar to the nominal output above, where the primary difference is
that each parameter set is nested one directory lower using the parameter set number.
$ ls build/mesh_convergence/
parameter_set0/ parameter_set1/ parameter_set2/ parameter_set3/
PS > Get-ChildItem build\mesh_convergence\
Directory: C:\Users\roppenheimer\waves-tutorials\waves_quickstart\build\mesh_convergence
Mode LastWriteTime Length Name
---- ------------- ------ ----
d---- 6/9/2023 4:32 PM parameter_set0
d---- 6/9/2023 4:32 PM parameter_set1
d---- 6/9/2023 4:32 PM parameter_set2
d---- 6/9/2023 4:32 PM parameter_set3
These set names are managed by the WAVES parameter study object, which is written to a separate build directory for later re-execution. This parameter study file is used in the parameter study object construction to identify previously created parameter sets on re-execution.
$ ls build/parameter_studies/
mesh_convergence.h5
$ waves print_study build/parameter_studies/mesh_convergence.h5
set_hash width height global_seed displacement
set_name
parameter_set0 cf0934b22f43400165bd3d34aa61013f 1.0 1.0 1.000 -0.01
parameter_set1 ee7d06f97e3dab5010007d57b2a4ee45 1.0 1.0 0.500 -0.01
parameter_set2 93de452cc9564a549338e87ad98e5288 1.0 1.0 0.250 -0.01
parameter_set3 49e34595c98442a228efd9e9765f61dd 1.0 1.0 0.125 -0.01
PS > Get-ChildItem build\parameter_studies\
Directory: C:\Users\roppenheimer\waves-tutorials\waves_quickstart\build\parameter_studies
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 9942 mesh_convergence.h5
PS > waves print_study build\parameter_studies\mesh_convergence.h5
set_hash width height global_seed displacement
set_name
parameter_set0 cf0934b22f43400165bd3d34aa61013f 1.0 1.0 1.000 -0.01
parameter_set1 ee7d06f97e3dab5010007d57b2a4ee45 1.0 1.0 0.500 -0.01
parameter_set2 93de452cc9564a549338e87ad98e5288 1.0 1.0 0.250 -0.01
parameter_set3 49e34595c98442a228efd9e9765f61dd 1.0 1.0 0.125 -0.01
Try adding a new global mesh seed in the middle of the existing range. An example might look
like the following, where the seed 0.4 is added between 0.5 and 0.25.
mesh_convergence_parameter_generator = waves.parameter_generators.CartesianProduct(
{
"width": [1.0],
"height": [1.0],
"global_seed": [1.0, 0.5, 0.4, 0.25, 0.125],
"displacement": [-0.01],
},
output_file=mesh_convergence_parameter_study_file,
previous_parameter_study=mesh_convergence_parameter_study_file
)
Then re-run the parameter study. The new parameter set should build under the name parameter_set4 and workflow
should build only parameter_set4 tasks. The parameter study file should also be updated in the build directory.
$ scons mesh_convergence --jobs=4
$ ls build/mesh_convergence/
parameter_set0/ parameter_set1/ parameter_set2/ parameter_set3/ parameter_set4/
$ waves print_study build/parameter_studies/mesh_convergence.h5
set_hash width height global_seed displacement
set_name
parameter_set0 cf0934b22f43400165bd3d34aa61013f 1.0 1.0 1.000 -0.01
parameter_set1 ee7d06f97e3dab5010007d57b2a4ee45 1.0 1.0 0.500 -0.01
parameter_set2 93de452cc9564a549338e87ad98e5288 1.0 1.0 0.250 -0.01
parameter_set3 49e34595c98442a228efd9e9765f61dd 1.0 1.0 0.125 -0.01
parameter_set4 4a49100665de0220143675c0d6626c50 1.0 1.0 0.400 -0.01
PS > scons mesh_convergence --jobs=4
PS > Get-ChildItem build\mesh_convergence\
Directory: C:\Users\oppenheimer\waves-tutorials\waves_quickstart\build\mesh_convergence
Mode LastWriteTime Length Name
---- ------------- ------ ----
d---- 6/9/2023 4:32 PM parameter_set0
d---- 6/9/2023 4:32 PM parameter_set1
d---- 6/9/2023 4:32 PM parameter_set2
d---- 6/9/2023 4:32 PM parameter_set3
d---- 6/9/2023 4:32 PM parameter_set4
PS > waves print_study build/parameter_studies/mesh_convergence.h5
set_hash width height global_seed displacement
set_name
parameter_set0 cf0934b22f43400165bd3d34aa61013f 1.0 1.0 1.000 -0.01
parameter_set1 ee7d06f97e3dab5010007d57b2a4ee45 1.0 1.0 0.500 -0.01
parameter_set2 4a49100665de0220143675c0d6626c50 1.0 1.0 0.400 -0.01
parameter_set3 93de452cc9564a549338e87ad98e5288 1.0 1.0 0.250 -0.01
parameter_set4 49e34595c98442a228efd9e9765f61dd 1.0 1.0 0.125 -0.01
If the parameter study naming convention were managed by hand it would likely be necessary to add the new seed to the
end of the parameter study to guarantee that the new set received a new set number. In practice, parameter studies are
often defined programmatically as a range() or vary more than one parameter, which makes it difficult to predict how
the set numbers may change. It would be tedious and error-prone to re-number the parameter sets such that the
input/output relationships are consistent. In the best case, mistakes in set re-numbering would result in unnecessary
re-execution of the previous parameter sets. In the worst case, mistakes could result in silent inconsistencies in
the input/output relationships and lead to errors in result interpretations.
WAVES first looks for and opens the previous parameter study file saved in the build directory, reads the previous set definitions if the file exists, and then merges the previous study definition with the current definition along the unique set contents identifier coordinate. Under the hood, this unique identifier is an md5 hash of a string representation of the set contents, which is robust between systems with identical machine precision. To avoid long, unreadable hashes in build system paths, the unique md5 hashes are tied to the more human readable set numbers seen in the mesh convergence build directory.
Output Files#
Explore the contents of the build directory, as shown below.
$ pwd
/home/roppenheimer/waves-tutorials/tutorial_gmsh
$ find build/nominal -type f
build/nominal/SConscript
build/nominal/post_processing.py
build/nominal/rectangle.py
build/nominal/rectangle_compression.01.vtu
build/nominal/rectangle_compression.02.vtu
build/nominal/rectangle_compression.03.vtu
build/nominal/rectangle_compression.04.vtu
build/nominal/rectangle_compression.05.vtu
build/nominal/rectangle_compression.06.vtu
build/nominal/rectangle_compression.07.vtu
build/nominal/rectangle_compression.08.vtu
build/nominal/rectangle_compression.09.vtu
build/nominal/rectangle_compression.10.vtu
build/nominal/rectangle_compression.12d
build/nominal/rectangle_compression.cvg
build/nominal/rectangle_compression.dat
build/nominal/rectangle_compression.frd
build/nominal/rectangle_compression.frd.stdout
build/nominal/rectangle_compression.h5
build/nominal/rectangle_compression.h5.stdout
build/nominal/rectangle_compression.inp
build/nominal/rectangle_compression.inp.in
build/nominal/rectangle_compression.pvd
build/nominal/rectangle_compression.sta
build/nominal/rectangle_compression.vtu.stdout
build/nominal/rectangle_gmsh.inp
build/nominal/rectangle_gmsh.inp.stdout
build/nominal/rectangle_mesh.inp
build/nominal/rectangle_mesh.inp.stdout
build/nominal/spooles.out
build/nominal/stress_strain.csv
build/nominal/stress_strain.pdf
build/nominal/stress_strain.pdf.stdout
build/nominal/stress_strain.png
build/nominal/stress_strain.png.stdout
build/nominal/strip_heading.py
build/nominal/time_points.inp
build/nominal/vtu2xarray.py
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh
PS > Get-ChildItem -Path build\nominal -Recurse -File
Directory: C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh\build\nominal
Mode LastWriteTime Length Name
---- ------------- ------ ----
-a--- 6/9/2023 4:32 PM 9250 post_processing.py
-a--- 6/9/2023 4:32 PM 5405 rectangle_compression.01.vtu
-a--- 6/9/2023 4:32 PM 5485 rectangle_compression.02.vtu
-a--- 6/9/2023 4:32 PM 5415 rectangle_compression.03.vtu
-a--- 6/9/2023 4:32 PM 5475 rectangle_compression.04.vtu
-a--- 6/9/2023 4:32 PM 5399 rectangle_compression.05.vtu
-a--- 6/9/2023 4:32 PM 5384 rectangle_compression.06.vtu
-a--- 6/9/2023 4:32 PM 5387 rectangle_compression.07.vtu
-a--- 6/9/2023 4:32 PM 5460 rectangle_compression.08.vtu
-a--- 6/9/2023 4:32 PM 5338 rectangle_compression.09.vtu
-a--- 6/9/2023 4:32 PM 5338 rectangle_compression.10.vtu
-a--- 6/9/2023 4:32 PM 255 rectangle_compression.12d
-a--- 6/9/2023 4:32 PM 1938 rectangle_compression.cvg
-a--- 6/9/2023 4:32 PM 0 rectangle_compression.dat
-a--- 6/9/2023 4:32 PM 24021 rectangle_compression.frd
-a--- 6/9/2023 4:32 PM 15815 rectangle_compression.frd.stdout
-a--- 6/9/2023 4:32 PM 19463 rectangle_compression.h5
-a--- 6/9/2023 4:32 PM 9040 rectangle_compression.h5.stdout
-a--- 6/9/2023 4:32 PM 489 rectangle_compression.inp
-a--- 6/9/2023 4:32 PM 498 rectangle_compression.inp.in
-a--- 6/9/2023 4:32 PM 783 rectangle_compression.pvd
-a--- 6/9/2023 4:32 PM 830 rectangle_compression.sta
-a--- 6/9/2023 4:32 PM 5388 rectangle_compression.vtu.stdout
-a--- 6/9/2023 4:32 PM 434 rectangle_gmsh.inp
-a--- 6/9/2023 4:32 PM 886 rectangle_gmsh.inp.stdout
-a--- 6/9/2023 4:32 PM 343 rectangle_mesh.inp
-a--- 6/9/2023 4:32 PM 252 rectangle_mesh.inp.stdout
-a--- 6/9/2023 4:32 PM 5818 rectangle.py
-a--- 6/9/2023 4:32 PM 2280 SConscript
-a--- 6/9/2023 4:32 PM 0 spooles.out
-a--- 6/9/2023 4:32 PM 1014 stress_strain.csv
-a--- 6/9/2023 4:32 PM 12951 stress_strain.pdf
-a--- 6/9/2023 4:32 PM 1055 stress_strain.pdf.stdout
-a--- 6/9/2023 4:32 PM 1206 strip_heading.py
-a--- 6/9/2023 4:32 PM 44 time_points.inp
-a--- 6/9/2023 4:32 PM 6079 vtu2xarray.py
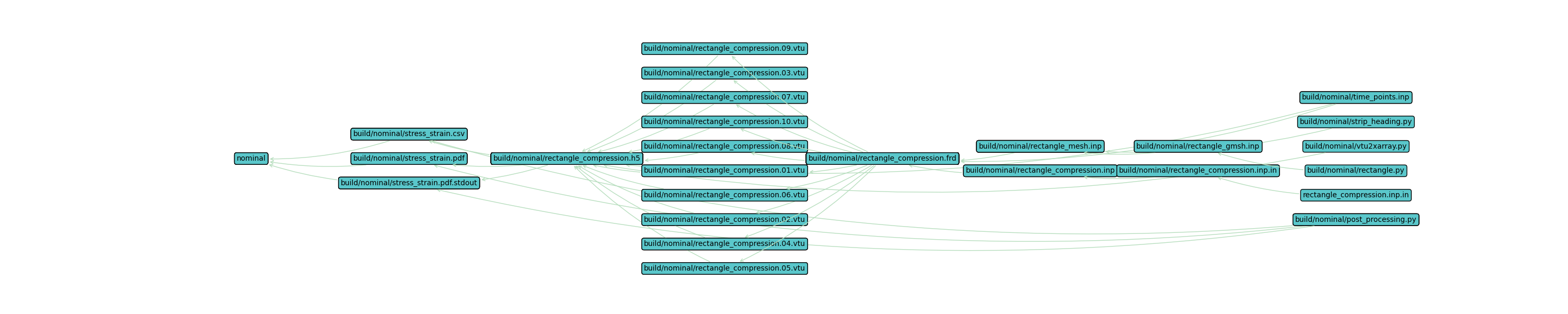
Workflow Visualization#
To visualize the workflow, you can use the WAVES visualize command. The --output-file allows
you to save the visualization file non-interactively. Without this option, you’ll enter an interactive matplotlib window.
$ pwd
/home/roppenheimer/waves-tutorials/tutorial_gmsh
$ waves visualize nominal --output-file tutorial_gmsh.png --width=30 --height=6
PS > Get-Location
Path
----
C:\Users\roppenheimer\waves-tutorials\tutorial_gmsh
PS > waves visualize nominal --output-file tutorial_gmsh.png --width=30 --height=6
The workflow visualization should look similar to the image below, which is a representation of the directed graph
constructed by SCons from the task definitions. The image starts with the final workflow target on the left, in this
case the nominal simulation target alias. Moving left to right, the files required to complete the workflow are
shown until we reach the original source file(s) on the far right of the image. The arrows represent actions and are
drawn from a required source to the produced target. The Computational Practices introduction discusses the
relationship of a Build System task and Directed graphs.